Molecular detection technology based on DNA variation is widely used in the fields of biomolecules, genomics, genetics, and breeding. In terms of animal and plant genetic breeding, DNA variation has been developed into different types of molecular markers such as SSR and SNP for genetic map construction, diversity analysis, marker-character association, map-based cloning, molecular breeding, fingerprint identification, etc. The development of various high-throughput detection equipment and high-density DNA chips greatly meets the needs of high-throughput and high-density molecular detection technology in animal and plant genetic breeding. However, chip development and production are difficult, and expensive testing equipment is required.
What We Offer
Lifeasible has successfully developed a high-resolution multi-SNP (multiple single-nucleotide-polymorphism cluster) detection system through targeted integration technology. This system can replace solid-phase chips and other molecular detection technologies that rely on expensive instruments, and is widely used in many fields such as animal and plant genetic breeding.
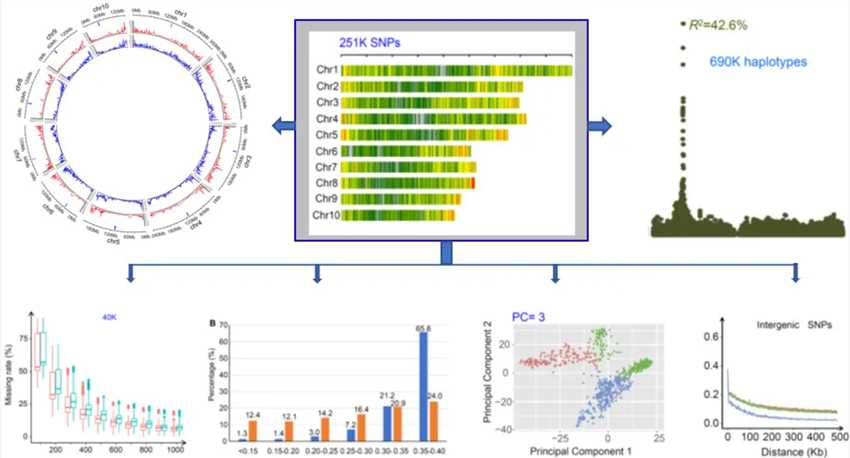 Figure 1. High resolution multi-SNP (mSNP) detection technology system and its application. (Guo Z, et al. 2021)
Figure 1. High resolution multi-SNP (mSNP) detection technology system and its application. (Guo Z, et al. 2021)
High-resolution Multi-SNP Technology Flow
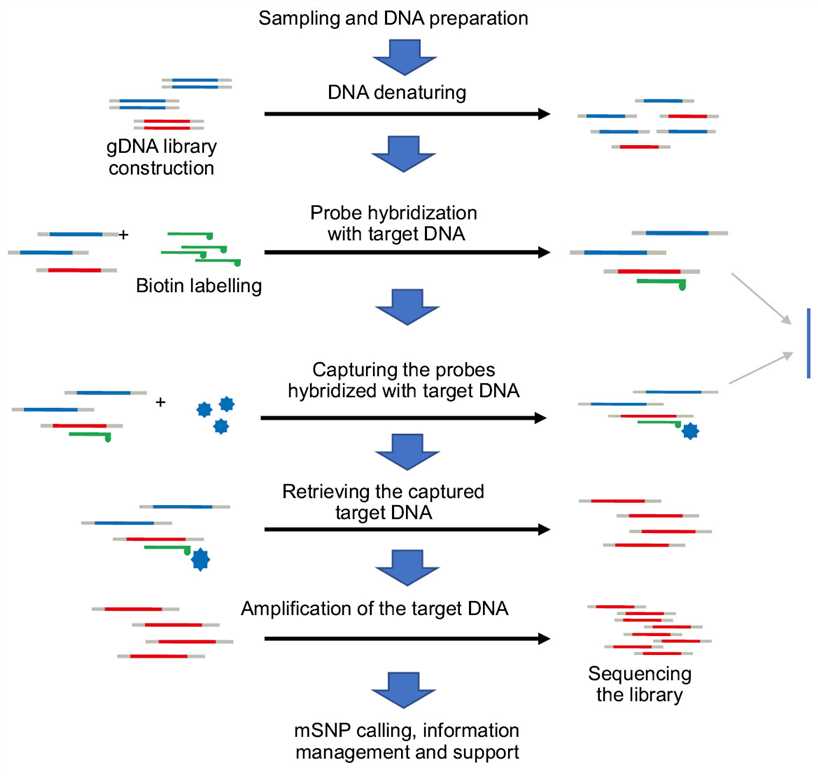 Figure 2. Technical procedure of genotyping by target sequencing (GBTS). (Guo Z, et al. 2021)
Figure 2. Technical procedure of genotyping by target sequencing (GBTS). (Guo Z, et al. 2021)
Technical Details
Based on the genotyping by target sequencing (GBTS) technology, our research team has established and optimized the mSNP technology system that uses a single amplicon to detect multiple SNP markers, greatly improve the detection efficiency of variation within the target site. At present, more than 50 sets of GBTS-based liquid-phase chips have been developed in 13 major crops, vegetables, and some animal and microbial species, and have been widely used in the above-mentioned related fields.
Through screening and optimization in maize, we developed a set of markers (40K mSNP) including 40K target sites. On average, 6.6 SNP markers can be detected per locus (amplicon), and multiple SNP markers at the same locus can constitute multiple haploid forms.
Therefore, the marker set contains three different types of markers as a whole, namely 40K mSNP, 260K SNP/Indel and 912K haplotype. Since this detection of mSNP sites and SNP markers is achieved through sequencing, the number of labeled sites that can be captured by sequencing is directly proportional to the sequencing depth. Therefore, according to the demand for label density in the application scenario, by controlling the sequencing depth, a variety of different label densities can be obtained from the same label set. Including the number of any sites (amplicons) of mSNP from 1K to 40K and different numbers of SNP markers and haplotypes derived therefrom.
Sample Preparation
Lifeasible has been deeply involved in molecular labeling method development technology for many years, and we have rich experience and technology accumulation. We believe our all-round services will surely satisfy our customers.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
July 13, 2024