Plant transposable elements (TEs) are segments of DNA that can move or replicate in the genome and can be integrated into new loci in the genome. Plant transposable elements have important implications for the composition, evolution, transmission, and gene expression of plant genomes. With the rapid development of sequencing technology in recent years, more and more genomic data of plants have been published, providing a good opportunity for the detection and annotation of transposable elements.
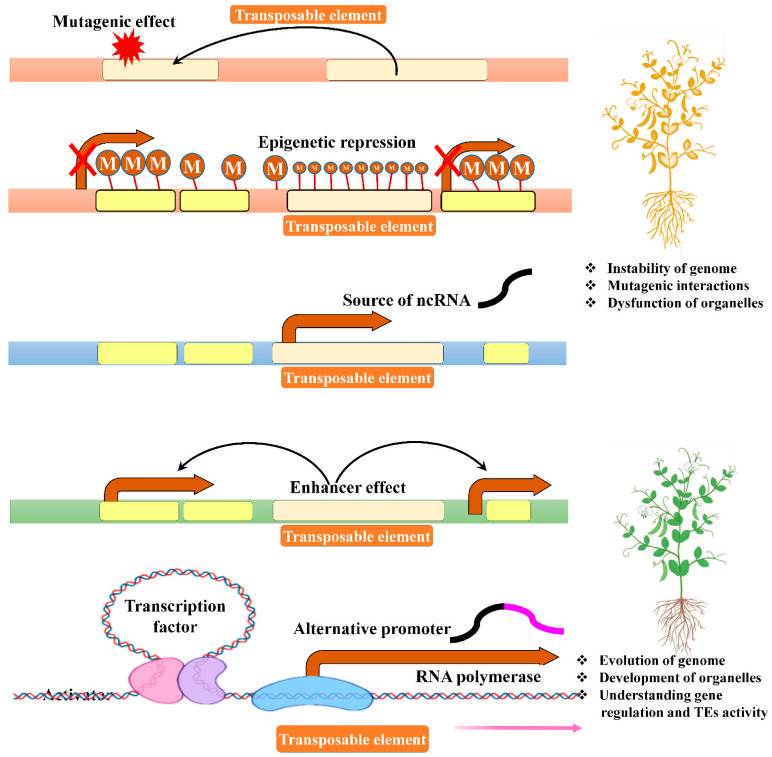 Figure 1. The major regulatory role of transposable factors in plants. (Ramakrishnan, M, et al. 2021)
Figure 1. The major regulatory role of transposable factors in plants. (Ramakrishnan, M, et al. 2021)
Lifeasible has a long history of research in the field of plant molecules, and our professional sequencing and bioinformatics platforms allow us to provide satisfactory identification and bioinformatics analysis of plant transposable elements and to analyze the sequences of transposable elements contained in plant genomes, with a view to understanding and exploiting transposable elements in plants comprehensively.
Sequencing of the plants to be analyzed is the first step in analyzing transposable elements. We will then perform sequence alignment of eukaryotic genomes using RepeatMasker and Greedier software, RepBase, and Dfam databases to annotate transposable elements homologously. The identified transposable elements will be family clustered, and their internal structural features will be resolved in detail using tools such as SSR Locator, TRF, IRF, and weblogo.
To analyze the effect of plant transposable elements on the structure and function of functional genes, we used the gene function clustering website agriGO, and the gene structure display website GSDS, to identify genes affected by insertions and to analyze the structure, function, and expression of these genes.
Transposable elements can enter the host through genomic hybridization, horizontal transfer, and other means, thereby affecting the expression of host genes. We explore potentially transcriptionally active transposable elements and analyze their effects on genotype and phenotype to help our clients understand the impact of transposable elements on plant gene expression.
Because the internal structure of some transposable elements is poorly conserved, we use their internally encoded transposase sequences for evolutionary analysis. After proofreading, the evolutionary tree of plant transposons is usually constructed using MEGA to analyze the evolutionary relationships of plant genomes.
Transposable elements play a vital role in different plants, and our powerful research tools allow us to study in detail the number, structural characteristics, chromosome distribution, insertion of genes, and expression patterns of transposable elements in plants, providing strong technical support for our client's research.

Lifeasible has accumulated extensive experience in transposable elements and can rapidly characterize transposable elements for our clients and scientifically predict and analyze the impact of transposable elements on plant gene evolution. If you have any questions, please do not hesitate to contact us.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
July 13, 2024