Nitrogen-fixing bacteria and clumping mycorrhizal fungi are responsible for the breakdown of most organic matter in the soil and provide essential nutrients to plants through a symbiotic relationship. This relationship can be enhanced by practices such as biodiversity enrichment and microbial fauna that occur in agroforestry-pastoral systems. As with many other plants, forage grasses have formed extensive associations with endosymbiotic heterotrophic microorganisms, including legumes such as T. repens (white clover) and M. sativa (alfalfa, zoysiagrass) with rhizobia. Currently, forage grasses can form relationships with clumping mycorrhizal fungi to improve phosphorus solubilization and uptake through additional strategies to cope with low nutrient availability. Forage grasses evolved with metabolites/nutrients that were important in controlling the exchange between partners.
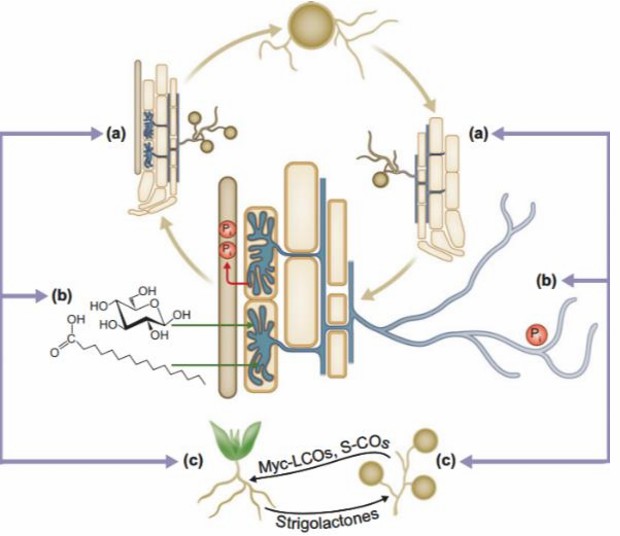 Fig.1. Interaction of soil microorganism in the rhizosphere (a) to acquire the soil nutrients (b) by plants (c). (Jardim A, et al., 2018)
Fig.1. Interaction of soil microorganism in the rhizosphere (a) to acquire the soil nutrients (b) by plants (c). (Jardim A, et al., 2018)
The effect of symbiotic forage plants on host adaptation usually depends on the symbiont or host genotype and environmental conditions. Primary metabolism undergoes significant changes during the transition from rhizosphere-free root tissue to functional rhizomes. Based on GC-MS and ultra-high performance LC-quadrupole time-of-flight-MS technology platforms, our experts use metabolic profiling or metabolomics to analyze metabolic changes in rhizobia and endophyte-infected forage plants.
As the ideal partner for forage metabolomics, Lifeasible offers customized solutions for the metabolomic analysis of symbiotic forage plants.
AMF plays a key role in acquiring inorganic phosphorus from insoluble phosphorus sources in the soil and transferring it to the host. We offer labeled isotope and NMR spectroscopy techniques to analyze the metabolomics of the forage grass-AMF association. Our goal is to help our clients identify fungus-specific metabolites, including alginans, unsaturated fatty acids, sterols, and carotenoids.
Endophytic bacteria produce a variety of alkaloids with anti-herbivory activity to protect host plants from herbivory by mammals, insects, and other invertebrates. We provide transcriptomic analyses of forage grasses in response to endophyte infection, reflecting metabolite exchange mechanisms by comparing targeted and untargeted metabolomic analyses of infected and uninfected plants.
In close collaboration with microbiologists, we are working to identify novel endophytic strains with beneficial metabolic profiles by using high-throughput analysis with direct MS injection to screen large numbers of infected seeds and forages rapidly. We can also achieve the creation of hybrid strains that exhibit improved species specificity in terms of symbiotic characteristics without the insertion of exogenous DNA.
Integrating modeling related to forage physiology and the efficacy of specific traits, we provide the most efficient metabolomics service for symbiotic forage plants to analyze interactions between forage genomes and their associated microbiota. In addition, we develop genetic manipulation strategies and strain combinations that mimic natural evolution to optimize plant-specific rhizobia inoculants for improved forage production. Lifeasible is one of the world's leading companies in forage metabolomics analysis. If you have any special requirements about our solutions, please feel free to contact us.
Reference
Lifeasible has established a one-stop service platform for plants. In addition to obtaining customized solutions for plant genetic engineering, customers can also conduct follow-up analysis and research on plants through our analysis platform. The analytical services we provide include but are not limited to the following:
July 13, 2024